bio_back_up_9_11_new
Topic outline
-
-
-
Use software tools designed to facilitate biodiversity data capture to produce digital biodiversity data from analogue sources.
Learn data quality concepts and receive an introduction to tools used for standardizing data, validating data, and cleaning data.
Use software tools to evaluate the fitness-for-use of a biodiversity dataset
Use software tools designed for (biodiversity) data cleaning.
Learn the process of making biodiversity data freely available online, also known as data publishing, utilizing GBIF’s Integrated Publishing Toolkit (IPT).
Define the publishable data types and subtypes (if any) for a biodiversity dataset.
Use the GBIF IPT to publish biodiversity datasets using the appropriate extensions.
Capacitate others in the planning, capture, management and publishing of biodiversity data.
-
Participants should possess the following skills and knowledge:
Basic skills in computer and internet use, and, in particular, in the use of spreadsheets.
Basic knowledge about geography and biodiversity informatics: geography and mapping concepts, basic taxonomy and nomenclature rules.
Willingness to disseminate the knowledge learned in the workshop with partners and collaborators in your project by adapting the biodiversity data mobilization training materials to specific contexts and languages while maintaining their instructional value.
A good command of English. While efforts are made to provide materials in other languages, instruction/videos will be in English.
-
-
-
-
 Here we will need a video introduction with the key messages of the projectThe science of biodiversity has accumulated probably one of the oldest and richest data pools on the living world, dating back to the ancient times and resulting in more than 500 million pages of published literature, more than 2 billion specimens in natural history collections and more than 1.8 million species described. A much greater volume of digital data will be generated in the coming years as we digitize the world’s biodiversity specimens, species observations, species traits, species names, classifications, and literature. To create actionable knowledge from this vast pool of data we need to link these digital objects together.
Here we will need a video introduction with the key messages of the projectThe science of biodiversity has accumulated probably one of the oldest and richest data pools on the living world, dating back to the ancient times and resulting in more than 500 million pages of published literature, more than 2 billion specimens in natural history collections and more than 1.8 million species described. A much greater volume of digital data will be generated in the coming years as we digitize the world’s biodiversity specimens, species observations, species traits, species names, classifications, and literature. To create actionable knowledge from this vast pool of data we need to link these digital objects together.Context of Use
Aims and Challenges
Structure of the BKH
-
On the top Menu of BKH the user can selct the FAIR DATA PLACE tab.
The following screen will appear:

This is the core component of BKH and presents FAIR data, services and tools developed during the BiCIKL project. In the future, the BKH will also accept services on linked data provided by new members and third parties.
The FAIR DATA PLACE consists of three sub-modules:
○ Research Infrastructures and Network Organisations: This sub-module lists the member organisations and research infrastructures with short descriptions and links to their data, tools and services.

Figure 1: In the FAIR DATA PLACE BKH users can explore Research Infrastructures and Network Organisations that are populating the services of the aggregator.
Users can explore (meta)data, tools and services offered by the participating Research Infrastructures and Network Organisations organised by category: Specimens, Sequences, Taxon names and Literature.
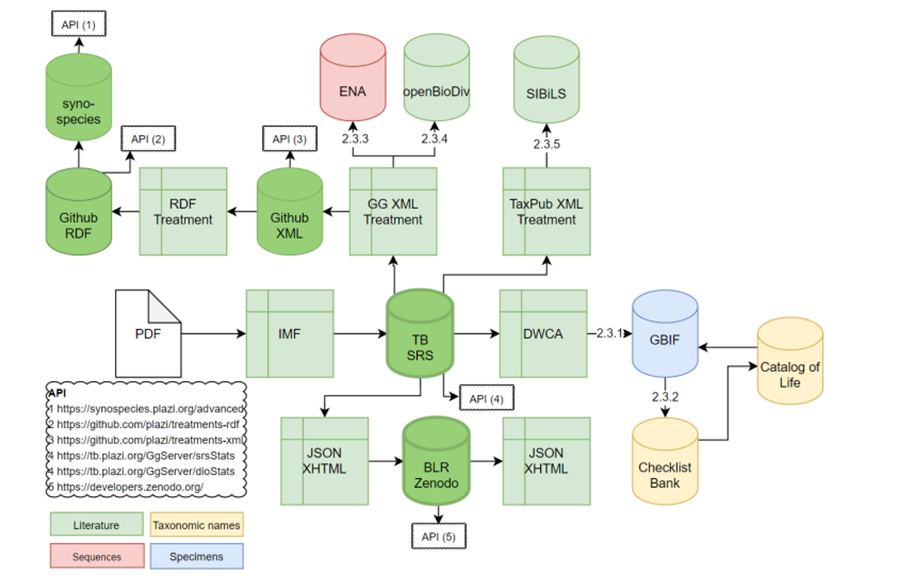
Figure 2. Overview of the data flow between literature based data and the data domains in BKH (specimens, taxonomic names, and sequences).

Figure 2: In the FAIR DATA PLACE BKH users can explore (meta)data, tools and services offered by the participating Research Infrastructures organised by category: Specimens, Sequences, Taxon names and Literature. Apart from the link the related repositories, BKH offers a short description of the related data types and the services of each provider.
○ Linked data services: A catalogue of novel services that deliver FAIR data linked between biodiversity research infrastructures. In the next section of this module of the course we will provide detailed guidance and support to the BKH users to explore these novel services.
○ Become a member application form (For Service Providers): A formal questionnaire which serves as a basis to check the suitability of an organisation or research infrastructure to join BKH. It has to be noted that before applying to become a contributor to BKH, please ensure that your RI is compliant with the FAIR data checklist criteria and interoperability guidelines.
-
The next component of BKH presents the Open Calls published by the BICIKL project. It provides guidelines and examples of good practice to help proposers to design and submit succesful proposals. Research projects that use linked FAIR biodiversity data are also presented. The page also displays a network analysis tool based on FAIRsharing.org data.
Open Call Projects
Network Analysis Tool
-
-
In this section we will introduce the key concepts related to the Data Liberation and the use of Publishing Tools available. More specifically we will discuss:
The current status of semantic publishing, how we can use it to provide enhanced data for addressing the current biodiversity crisis.
How natural history institutions can use semantic publishing and data outputs to help build their research strategies.
Showcase the exciting new semantic publishing tools currently in place and the status quo of moving from Open Access to Open Access FAIR data.
Linking between publications and collection-related data.
Linking external data resources and the role of publications as the seeds for building knowledge graph of BKH.
Presentation of two sets of semantic publishing tools: Plazi workflow and Pensoft ARPHA.
Lets start by presenting a graphical representation of the evolution and the current status of Publishing (2023).
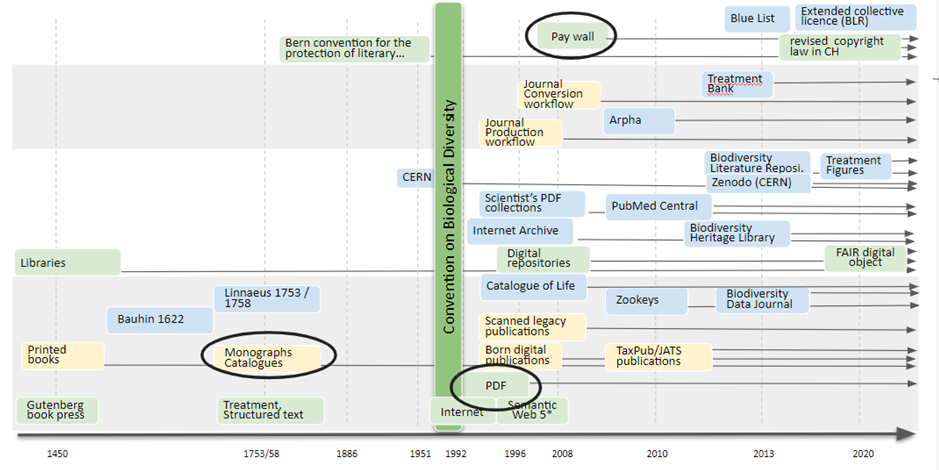
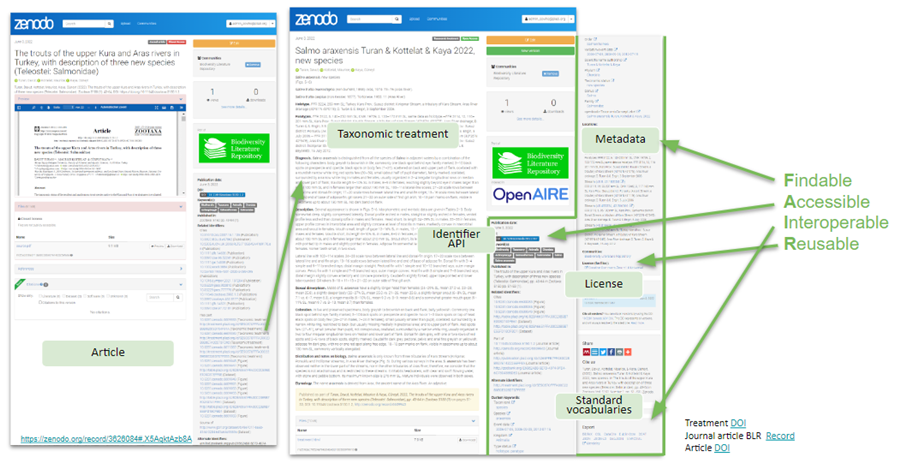
A key concept of the Digital Accessible Knowledge in the Taxonomic Treatment.
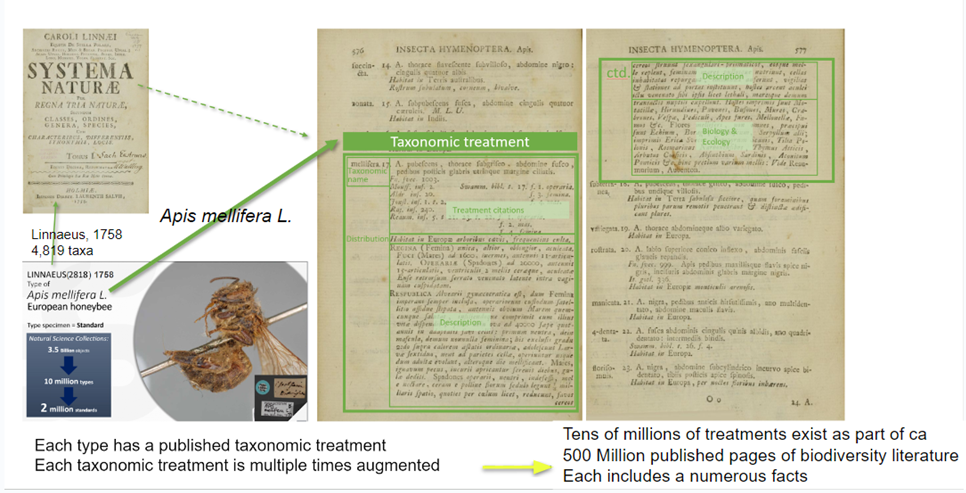
The following image presents Treatments as FAIR digital objects in the Biodiversity Literature Repository.
-
-
This section describes the key functionalities of the eBioDiv Matching Service that is used for linking material citations to specimens.
To facilitate the task, a semi-automatic matching approach is used. Users are presented with lists of possible matches. Once a sufficient number of matching decisions have been recorded by the users of the eBioDiv Matching Service, it will be possible to start exploring ways of gradually replacing the current semi-automatic matching approach by a fully-automatic matching algorithm for data pairs with a sufficient number of data points of score high enough to warrant automatic acceptance of a match.
Furthermore, it might become possible to automatically sort out data entries that are clearly deficient and therefore unfit as an input for matching decisions. These data entries could then be relegated back to the maintainers of the data along with a request to improve the data.
As a result, fewer matching suggestions will have to be curated by humans. It will be interesting to observe how the intellectual work carried out by the users of the eBioDiv Matching Service will evolve over time as a result of this shift in the balance between the tasks relegated to computer algorithms and the tasks remaining with human contributors.
If you want to contribute to the realization of this vision please refer to the following documentation to get an introduction to the matching process and to get access to detailed documentation related to the use of the Matching Service.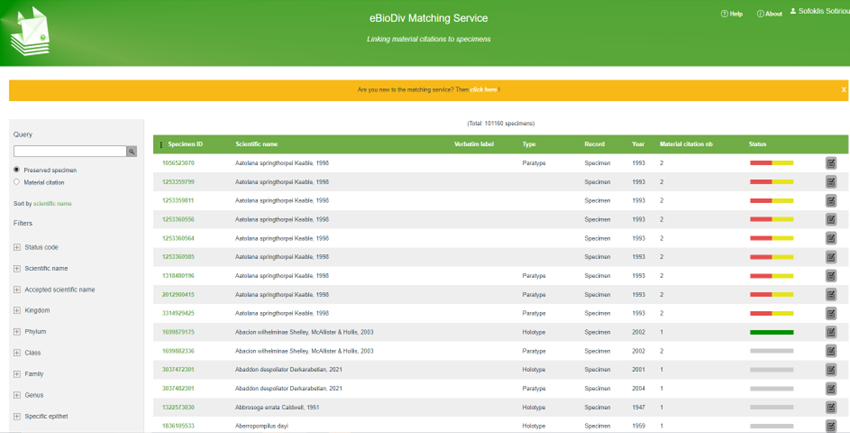
Figure 1. The eBioDiv Matching Service Interface.
- For new users it is highly recommended to have a close look to the following documentation to get an introduction to the matching process:
Introduction to the Matching Service
- For advanced users the key steps of the process are described in the following documentation:
-
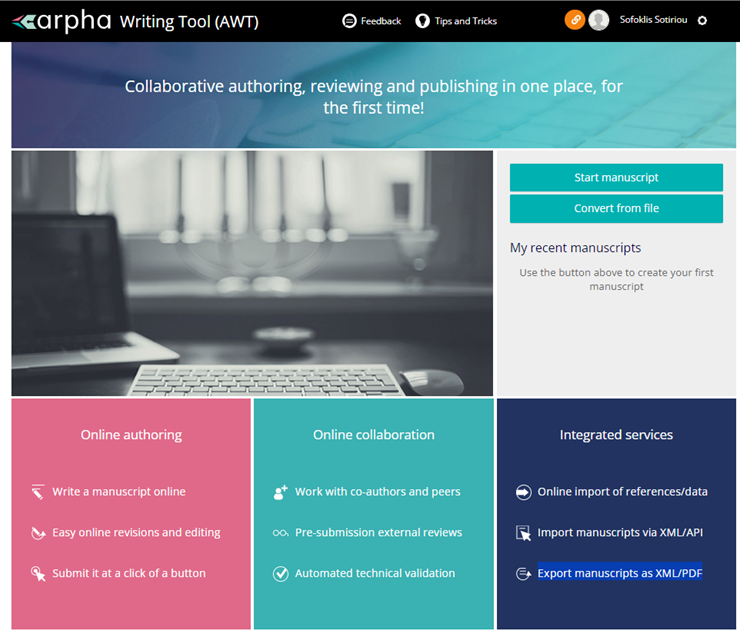
This Section presents the ARPHA Writing Tool. It offers a
one-stop entry and unified interface;
reduced manuscript turnaround times;
monitoring tools at all stages of the publishing process;
data security and GDPR compliance;
- optimised cost/quality ratio.
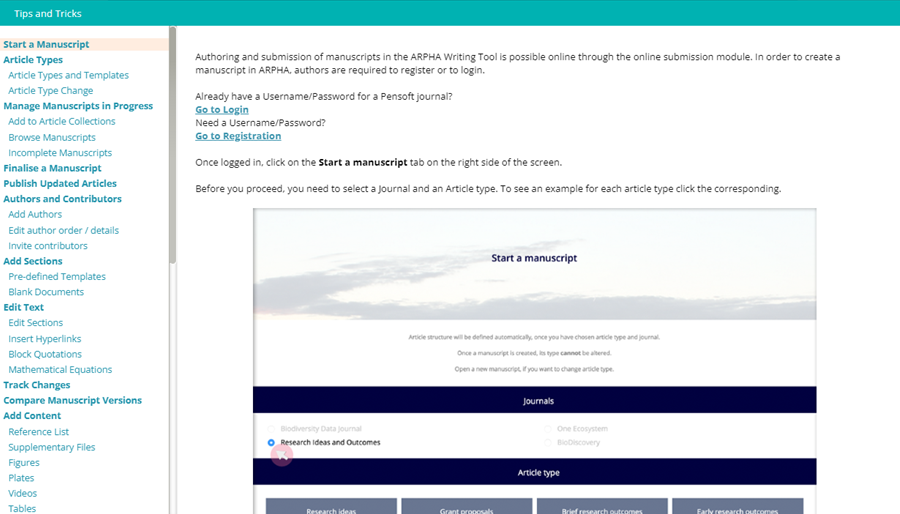
The Tips and Tricks Button offers a detailed manula of the service.
-
-
-


