Workbench
The Workbench offers two services, the GoldenGATE Imagine and the Matching Service. In the following section we are presenting in short their key role and functionalities in the overall process.
GoldenGATE Imagine
Data automatically liberated from literature can be annotated using GoldenGATE Imagine. This also allows to curate and attribute annotations like “accession code”, “collection code”, “specimen code” or “taxonomic name” with the respective identifiers. The GoldenGATE Document Editor is a visual editor for marking up documents. The main goal of the Imagine version is to provide a markup tool for digital born PDFs. It is designed to do most of the markup automatically; manual work is reduced to correcting the output of automated components. Accession numbers, specimen codes or collection codes can automatically be detected and attributed with the respective PIDs. However, there is no standard way to publish and compose these codes or numbers yet, and thus automated annotation needs human curation, which is enabled using the GGI UI (Fig. 1). Access to new documents or editing existing annotations requires authentication. Enhanced publications are versioned and can be rolled back to previous versions and assigned to a specific user (Fig. 2).
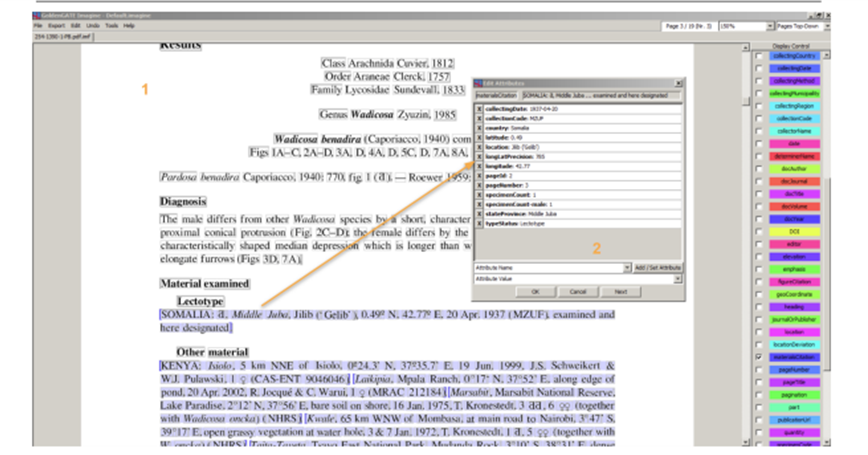
Figure 1: GoldenGate Imagine editor User Interface. 1 Main panel; 2 annotation editor
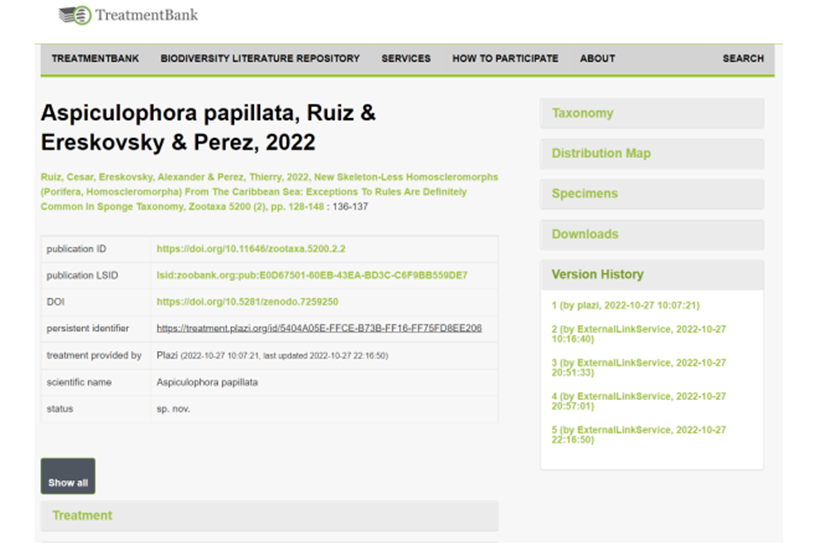
Figure 2: Treatment view in TreatmentBank showing the version history.
The Matching service is a dedicated tool to link material citations to preserved specimens in GBIF. For this purpose, GBIF is providing all the clusters of record relations it detects, including material citations, to TreatmentBank where, for each material citation, one or more matches are provided.
Material citations in publications traditionally cite a single preserved specimen, such as a holotype or groups of specimens. Material citations as part of a treatment provide scientific results based on the respective specimen, and in other words, add the specimen to the biodiversity knowledge graph. Today, most of the digital specimens are not accessible via APIs at their natural history museums, but the datasets are often published in GBIF. This provides an opportunity to use a single consistent matching service instead of developing individual solutions. The Matching service is based on the clustered occurrence data in GBIF, namely clusters including material citations from Plazi as well as occurrence records that represent museum specimens. In reality, the data have different codes, abbreviations, and omissions, so detailed match-up is based upon a one-by-one comparison of individual attributes of specimens that are cited in material citations, yielding per-attribute and overall matching scores. Once a curator or taxonomer has confirmed a match between a material citation and a museum specimen by inspecting the field comparison in the respective GUI (Fig. 3), that relationship is stored in the matching service internally, which then forwards the GBIF occurrence key of the specimen to TreatmentBank (via a dedicated REST endpoint / microservice), where it is written into the treatments, specifically into the material citation, and forwarded to GBIF when the DwCA is next published. Currently, the link to the GBIF occurrence is recorded in the DwC field “reference” in the occurrence.txt file. As an alternative to the GBIF occurrence key, any PID of a matching specimen can be added.
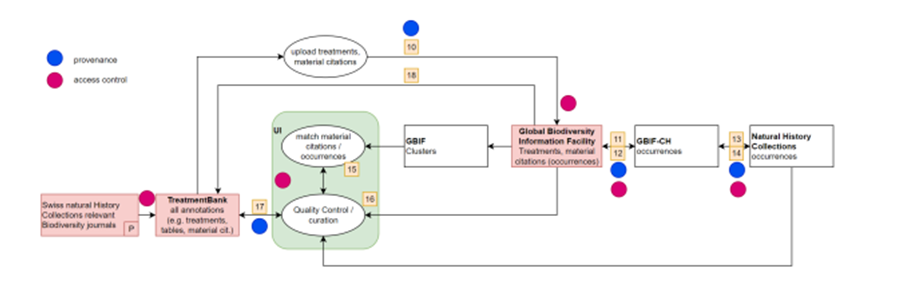
Figure 3: Schematic Matching service workflow.
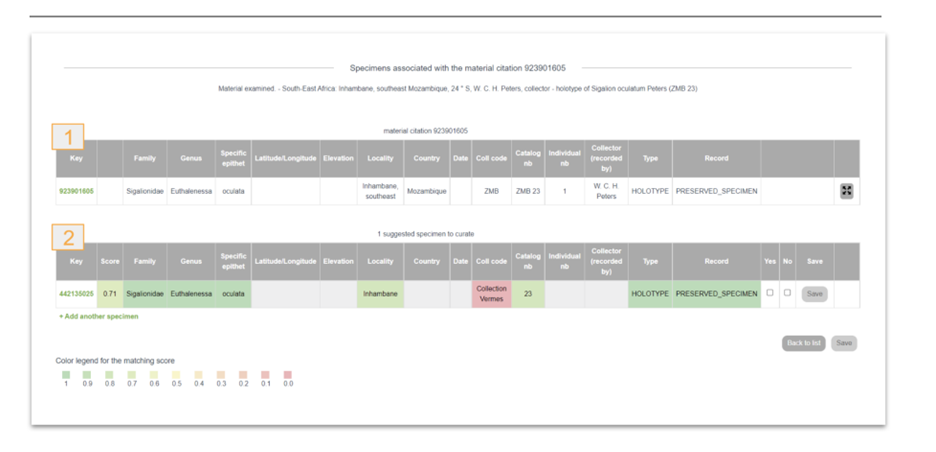
Figure 4: GUI displaying the match of a material citation reused by GBIF with a preserved specimen. Note the variety in the data fields. 1: material citation GBIF key; 2: preserved specimen GBIF key.