Introduction
The SIBiLS SPARQL Endpoint is a web service that allows users to query biomedical literature data using SPARQL queries. This can be particularly useful for researchers and professionals in the biomedical field who are looking for specific information within large datasets of biomedical literature.
You will find materials and resources to test the OpenLink Virtuoso SPARQL protocol endpoint in the Virtuoso Wiki page describing the service or in the RDF Data Access and Data Management chapter of the Online Virtuoso Documentation.
There is also a rich Web-based user interface with sample queries.
To use it, you have to ask the site admin to install the iSPARQL package (isparql_dav.vad). The SIBiLS SPARQL Endpoint can be used in various ways depending on the specific needs of the researcher. Running SPARQL queries on a subset of annotated biodiversity publications in PMC is a specialized task that requires both knowledge of SPARQL and an understanding of the specific annotations and data structure used in PMC for biodiversity-related publications. Here's a step-by-step example to illustrate how you might approach this:We can envision a research project where a team is interested in gathering and analyzing specific data. Here's a step-by-step breakdown of this scenario:
The research team aims to study the impact of climate change on a particular group of species, such as amphibians. They want to gather information from PMC publications that discuss climate change effects on amphibians, focusing on articles published in the last five years.
Preparation
- Familiarize with PMC Data Structure: The team first understands the structure of PMC data, including how articles are annotated and categorized.
- Determine Relevant Keywords: They decide on keywords like "amphibians", "climate change", "global warming", and specific species names.
Basic Query Structure: They construct a SPARQL query to fetch articles with these keywords in their abstracts or titles. The query also filters articles based on publication dates to focus on recent studies.
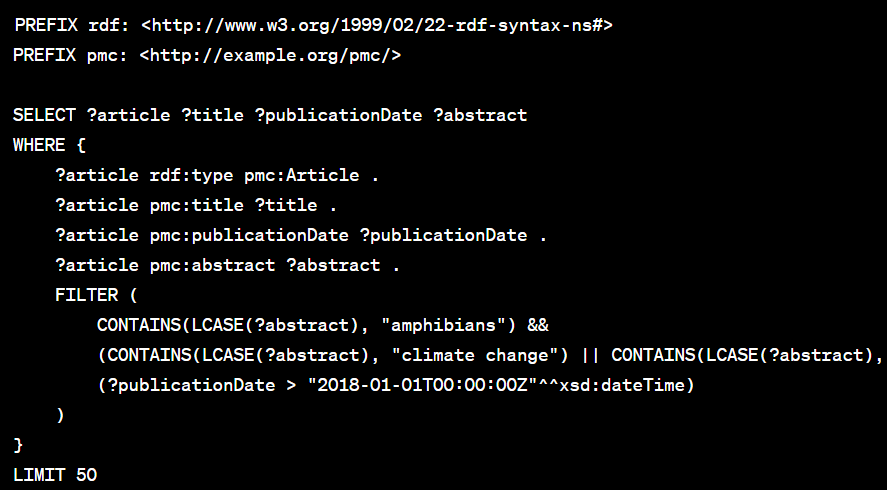
Running the Query
- Accessing the SIBiLS SPARQL Endpoint: They use the SIBiLS SPARQL Endpoint to run the query.
- Executing and Refining: The query is executed, and results are analyzed. If necessary, they refine the query for more precise results or to broaden the search.
- Collecting Data: The relevant articles' metadata (titles, abstracts, publication dates) are collected.
- Further Analysis: They may perform content analysis on the abstracts to identify common themes, trends, and gaps in the research.
- Deep Dive into Specific Articles: Based on initial findings, they might focus on specific articles for a more detailed review.
- Cross-referencing External Databases: They may also cross-reference their findings with external biodiversity databases or climate change datasets for a more comprehensive analysis.
- Compiling Findings: The team compiles their findings into a research report or paper.
- Sharing Data: They might also share their SPARQL queries and data extraction methods for reproducibility and to aid other researchers in similar studies.
- Monitoring Trends: They set up a system to periodically run these queries to monitor new publications in this domain.
- Expanding Research: Based on initial findings, the project might expand to include other species or environmental factors.
Create and Execute Queries Effectively
Running SPARQL queries, especially on complex datasets like those in PMC, can be challenging. Here are some tips to help you create and execute these queries more effectively:1. Understand the Data Schema
- Know the Structure: Familiarize yourself with the structure of the PMC data. Understanding how the data is organized (e.g., articles, authors, publication dates) is crucial for writing effective queries.
- Available Properties: Identify the available properties and how they are annotated in the dataset. This knowledge will help you access the correct information.
2. Define Clear Objectives
- Specific Goals: Be clear about what information you want to retrieve. This will help in constructing focused and efficient queries.
- Keywords and Filters: Decide on keywords and necessary filters (like date ranges, article types, etc.) in advance.
3. Optimize Query Performance
- Limit Results: Use the
LIMITclause to control the number of results, which is especially useful for testing and debugging. - Efficient Filtering: Use filters wisely. Overly complex filtering can slow down your query. Try to balance between precision and performance.
- Avoid Overly Broad Queries: Queries that are too broad may take a long time to execute and return more data than needed.
4. Test and Iterate
- Iterative Approach: Start with a simple query and gradually add complexity. This approach helps in isolating and fixing errors.
- Check Results: Regularly check if the results match your expectations. It's easier to spot issues in smaller result sets.
5. Use SPARQL Features Effectively
- Regular Expressions: Utilize SPARQL’s regular expression capabilities for more sophisticated text matching.
- Optional Patterns: Use
OPTIONALto include data that might not be present in all records, thereby avoiding missing out on partially matching records.
6. Handle Data Variability
- Case Sensitivity: Be mindful of case sensitivity. Use functions like
LCASE()to standardize text for comparison. - Data Inconsistencies: Be prepared to handle inconsistencies or unexpected formats in the data.
7. Leverage Sub-queries and Joins
- Sub-queries: Use sub-queries to break down complex queries into manageable parts.
- Joins: Understand how to effectively use joins to combine data from different parts of the dataset.
8. Utilize Available Tools and Documentation
- SPARQL Endpoints: Use the SPARQL endpoint's interface effectively. Some interfaces offer helpful features like query validation and formatting.
- Documentation: Refer to PMC’s documentation for specific details about their data structure and any custom SPARQL functions they might offer.
9. Plan for Query Evolution
- Scalability: Design queries that can be easily modified or expanded as your research needs evolve.
- Documentation: Keep a record of your queries and any changes you make, along with comments explaining the purpose and functionality of each part of the query.
10. Community and Forums
- Seek Help: If you encounter issues, don't hesitate to seek help from online communities or forums where SPARQL and PMC data experts congregate.
By following these tips, you can write more effective SPARQL queries for PMC biodiversity data, leading to more accurate and useful results for your research.
Endpoint Information
Consult the following links for available information defined on this endpoint:
What is SPARQL?
SPARQL is the W3C"s declarative query language for Graph Model Databases and Stores.
As is the case with SQL for relational databases and XQUERY for XML databases, SPARQL is an independent database and host operating system.
The development and evolution of this standard is overseen by the SPARQL Working Group within W3C. While parts of the language are still in active development, it is fully documented and publicly available.